The goal of this, or any other clustering tool on offer, is to identify groups of people that likely descended from a common ancestor. Those potential groups can be identified by their colour and placement on the chart.
A short guide to reading a matrix chart
- Names of my matches are listed down the left-hand side and repeated along the top of the matrix (I've blurred them for privacy).
- If there is a filled block at the intersection of two names (one at the side and one at the top) then those two people are a shared match.
- Coloured blocks indicate clusters (as defined by the algorithm used).
- Some people have connections to more than one cluster. Look to the grey blocks to see where those linkages are.
- If there are a lot of grey blocks between two clusters, then those clusters are probably relevant to each other. For example, the first (red) and fourth (green) groups have several connections between several people.
First impressions
The first thing I noticed was that the matrix was very fragmented. That could be representative of my data, but having browsed my DNA matches all those very small groups didn't feel quite right.I liked the amount of information given for the thresholds used:
"Your AutoCluster analysis was generated using thresholds of 25 cM (minimum) and 350 cM (maximum). In addition, DNA Matches were required to share at least 15 cM with one another in order to be indicated with a colored or gray cell. A total number of 104 DNA Matches ended up in 26 clusters in the final analysis."
Matrix visualisations are limited in how many matches they can include in one view and still be readable. Filtering is necessary to limit the matches. The automatically selected thresholds seem reasonable.
I appreciated the list of 11 matches who had no shared matches at the thresholds used.
I was perturbed by the exclusion of 95 matches who both met the threshold and had shared matches:
"The following 95 matches met the inclusion criteria but ended up in singleton clusters without other members and are therefore excluded from the analysis as well."95 matches in "singleton clusters"?! Why are there almost as many matches excluded for "singleton clusters" as there are matches actually included in the matrix? Just how aggressively does the algorithm chop up the groups?
As I read the long list of matches that had been excluded I was taken aback to see that my second closest match, at 129 cM shared, was among the "singleton clusters".
Digging deeper: A new view
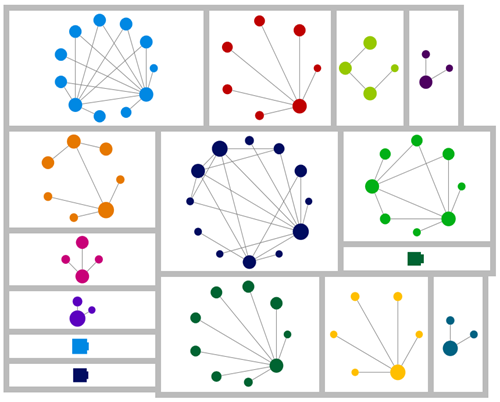
If you've been following my blog, you'll know that network graphs are my favoured tool for understanding shared match relationships. Using the csv file provided with the output, I was able to wrangle the data into shape and create a network graph version of the AutoCluster matrix information.I've aligned the group labels and colours with the matrix display (but made the second and third use of each colour darker for clarity). The numbers indicate the group in the AutoCluster result reading down the diagonal. The dot sizes reflect the amount of DNA I share with each person. Each line is a shared match relationship (the lines here are equivalent to the blocks in the matrix chart).
This was the result.
Looking at this graph, I retain my first impression that the algorithm is heavy-handed in breaking up the groups. For example, groups 1, 4 and two elements of thirteen look like they should be together, as do 24-25, and 3-19-22.
I'm relaxed about which group the closer match in group 11 is allocated to. That person would naturally "belong" in more than one cluster as they would likely match with groups of people with more distant ancestors from each side of our shared branch.
With this view, I also see that there are a few 'strings' of small groups. They include matches for whom, without more information, inclusion in one group or the next would be equally valid. That can't be helped when working with shared match information alone but is a reason to take care when looking at small groups in a matrix layout and track back to any other connected groups.
There is huge potential for refinement of matching groups with the data MyHeritage has - and that I'd like to get hold of as downloads! Information about the total size of the match between pairs, and whether the matches have a triangulated segment could be very informative to group allocations.
How would this have looked if the other 95 matches were included? I suspect that the sensible breakup of some of the smaller groups would be clearer for a start.
Digging even deeper - segment data
One of the website tools that I like on my MyHeritage is the chromosome browser tool. I entered the names of matches in my proposed larger group into the tool in batches. I both started and ended with my closest match. The result was clear. All of the matches I identified had triangulated segments with me and each other on chromosome three. (I couldn't find one match from group 4 in my match list to make that comparison).
Conclusions
I have only reviewed my own results and they may not be typical of most users. There seems to be an overly aggressive breakup of groups. This has made the chart fragmented and harder to read and interpret than it otherwise would be.The excessive fragmentation of groups is also likely the reason that almost half of my relevant matches were assigned to "singleton clusters" and excluded. Some of my best and most useful matches have been excluded. I'm concerned that the baby has been thrown out with the bathwater here.
When using AutoClusters I would suggest that users should:
- Read the notes. Take note of who's in and out.
- Use the grey cells to check for connections between groups.
- Don't assume that the matrix will include your best and closest matches. They could be excluded!
Overall though my feeling about the AutoCluster tool is that something is better than nothing. The AutoCluster tool is a helpful way to start identifying groups at the top end of your match list, but caution is needed.